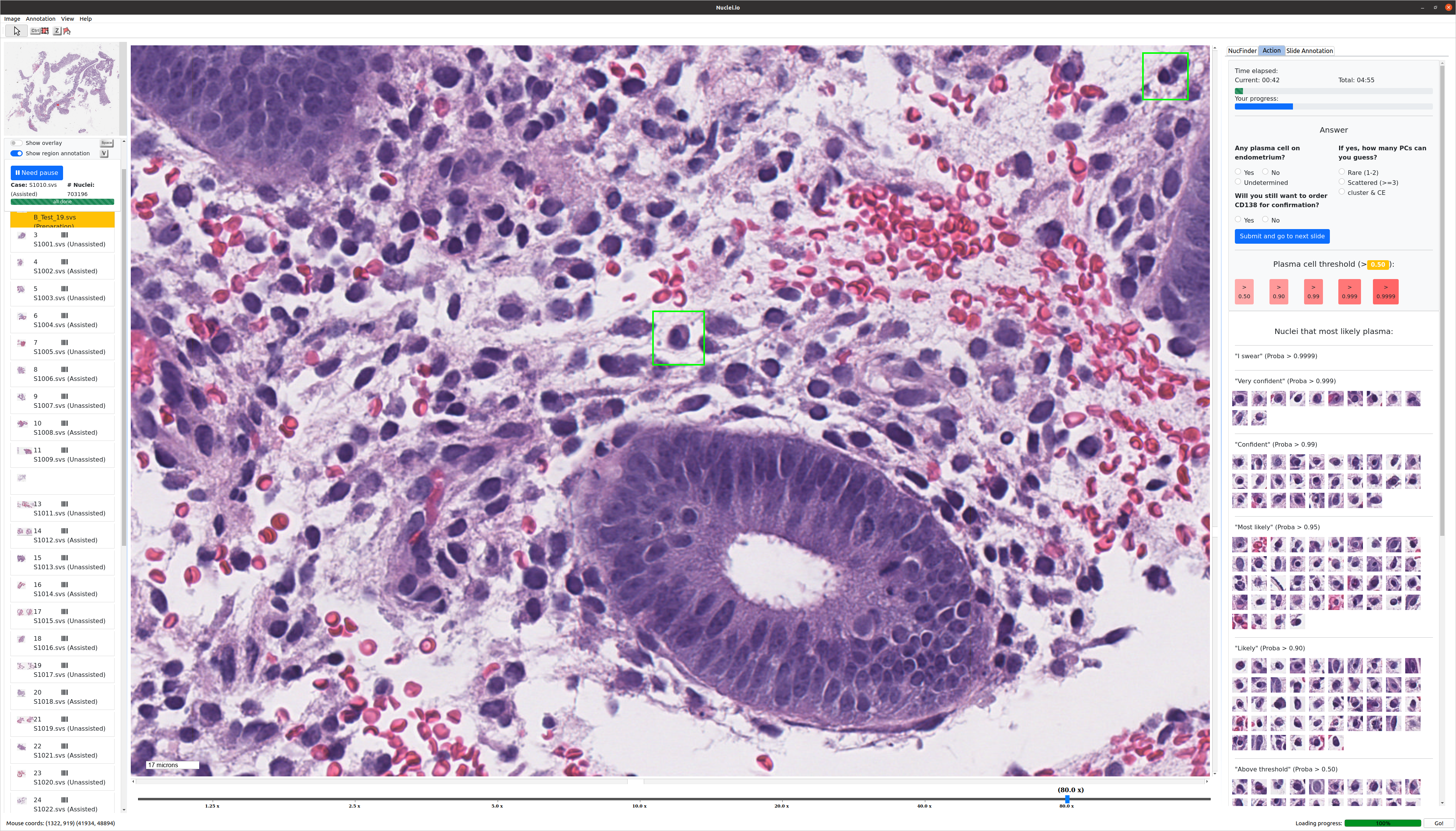
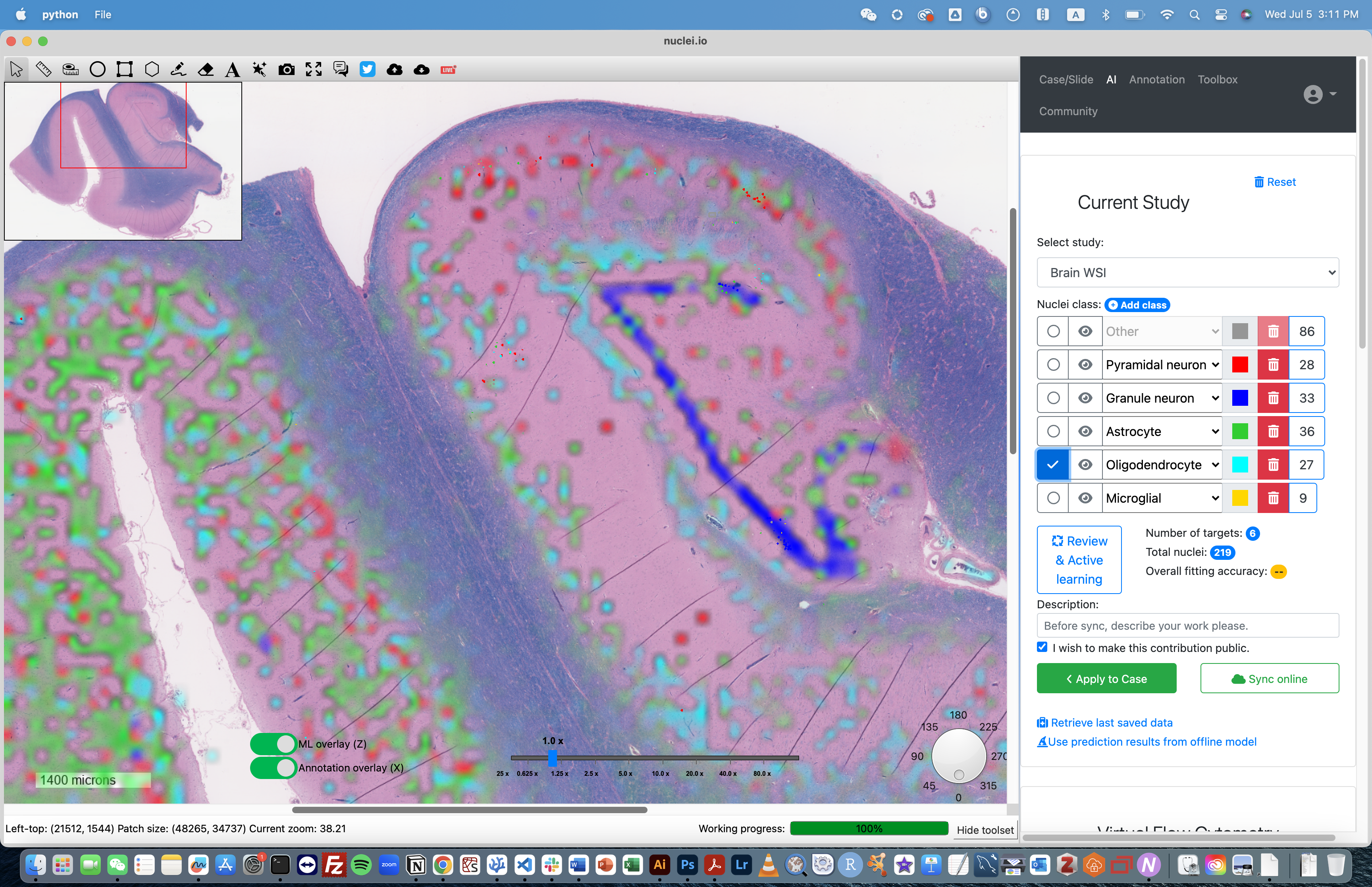
Project Highlights

- Pathologist-AI collaboration framework.
- Active learning with human-in-the-loop.
- Rapidly create a dataset in real-time.
- Faster and Better diagnosis.
Takeaway message
Nuclei.io enables pathologists to rapidly create diverse datasets and models for multiple clinically critical applications. To validate the effectiveness of this framework, we perform two cross-over user studies using a pathologist–AI collaborative approach. In both studies, the use of AI yielded considerable improvements in sensitivity, certainty, and efficiency in pathological diagnosis. Our findings validate the proposed human-in-the-loop framework, and suggest that pathologist–AI collaboration holds the potential to revolutionize digital pathology, ultimately leading to improved quality, efficiency, and cost.